In this tutorial we show you how to query interactions from the intercellular interactions in OmniPath, and go over the various attributes accompanying them.
We'll start by importing libraries, first omnipath,
and pandas for data wrangling.
import omnipath as op
import pandas as pd
The intercellular interactions in OmniPath are collated from a number of sources. When putting together a query, you can select all of these, or just a preferred subset of them. The requests.Intercell.resources function returns the list of datasets included in the database.
op.requests.Intercell.resources()
These resources contain a large variety of actors we can use to build intercellular interactions. Take a peek at a generalized list of these categories by using the requests.Intercell.generic_categories() function.
This list is also accessible from the browser, at https://omnipathdb.org/intercell_summary.
Using the requests.Intercell.categories() command returns the complete list.
op.requests.Intercell.generic_categories()
Now that we have seen the resources and categories, we have to go over a few definitions related to them to make sure everything is clear going forward.
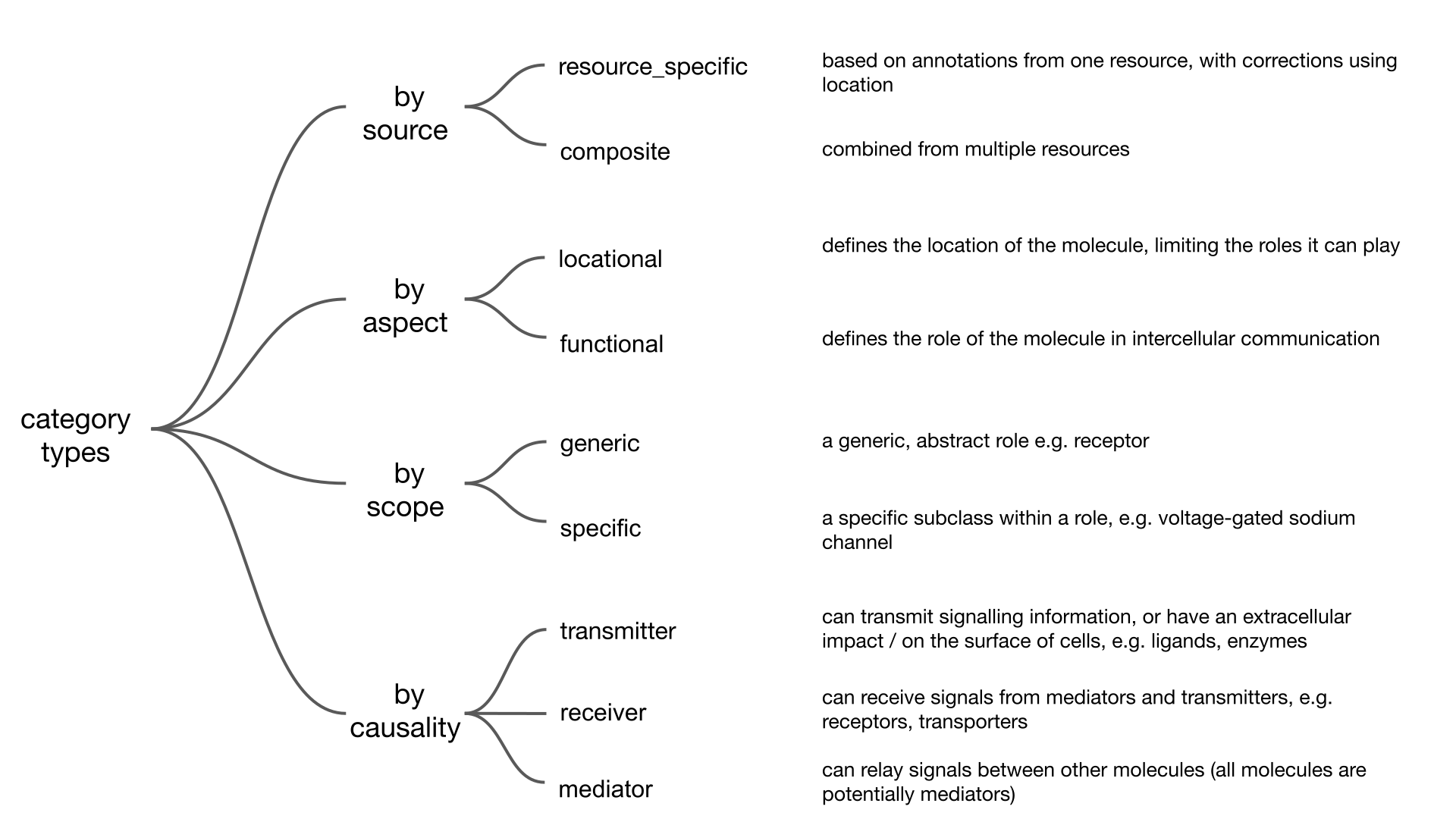
To import an intercellular network we call the interactions.import_intercell_network() function.
In this example below we generate a large intercellular network, where we are attempting to connect ligands to receptors.
- first, we define the datasets to import from
include - following that, we will specify the qualities of the transmitting proteins, such as which categories they should belong to, what should be their scope, source, aspect and so on under
transmitter_param - once we are done with that, we should specify the same for the receiving molecules under
receiver_param
These steps can be individually traced back through URLs:
- interaction parameters: https://omnipathdb.org/interactions?genesymbols=yes&datasets=omnipath,pathwayextra,ligrecextra&organisms=9606&fields=sources,references,curation_effort&license=academic
- transmitter parameters: https://omnipathdb.org/intercell?scope=generic&categories=ligand&causality=trans&license=academic
- receiver parameters: https://omnipathdb.org/intercell?scope=generic&categories=receptor&causality=rec&license=academic
intercell = op.interactions.import_intercell_network(
include=['omnipath', 'pathwayextra', 'ligrecextra'] # define categories
)
intercell_filtered = intercell[
(intercell['category_intercell_source'] == 'ligand') & # set transmitters to be ligands
(intercell['category_intercell_target'] == 'receptor') # set receivers to be receptors
]
intercell_filtered
This results in 7604 interactions. Let's narrow it down by restricting it with some of the categorical data outlined above.
intercell_filtered = intercell_filtered[
(intercell_filtered['category_source_intercell_target'] == 'resource_specific')
]
intercell_filtered
In this tutorial we learned:
- the data sources used to generate intercellular interactions
- the qualities of intercellular interactors
- the functions to generate and specify intercellular interactions